Resources
Frequently Asked Questions (FAQ)
Q: What sequencing platforms do you offer?
A: We carry Illumina and PacBio.
Q: Do you offer Nanopore or MGI sequencing?
A: Not at the moment.
Q: What’s the difference between Illumina and PacBio sequencing?
A: Illumina is based on short-reads sequencing technology while PacBio is based on long-reads sequencing technology.
Q: What is the standard or recommended sequencing depth for targeted metagenomics?
A: Typically 50,000 or 100,000 or 200,000 reads (Illumina platform) per sample is recommended for universal primers targeting 16S, 18S, ITS genes.
Q: What is the standard or recommended sequencing depth for shotgun metagenomics?
A: Usually 10 GB of data output is advised. However, the data output is also open for discussion based on the project needs.
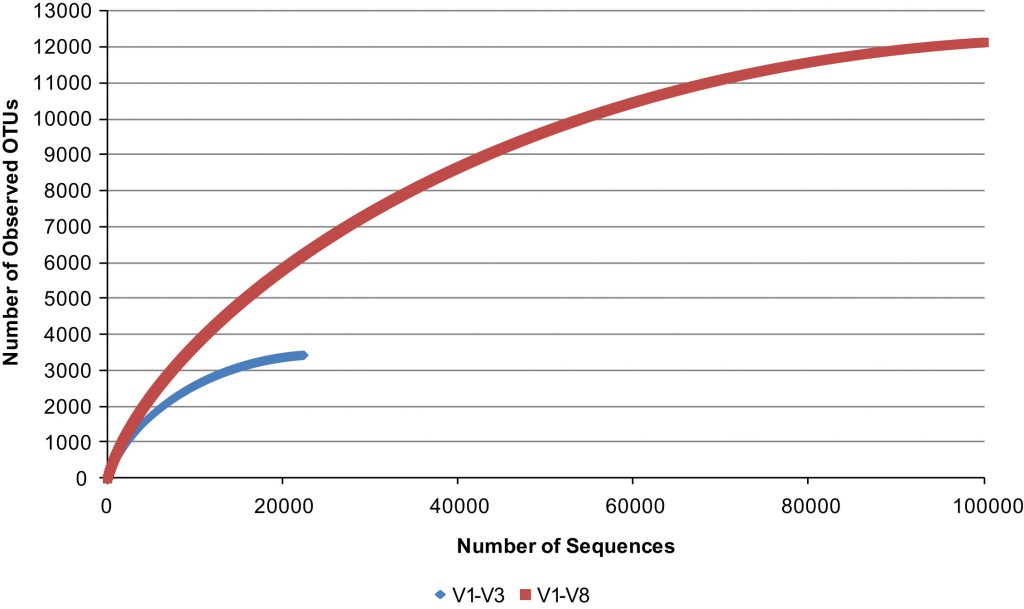
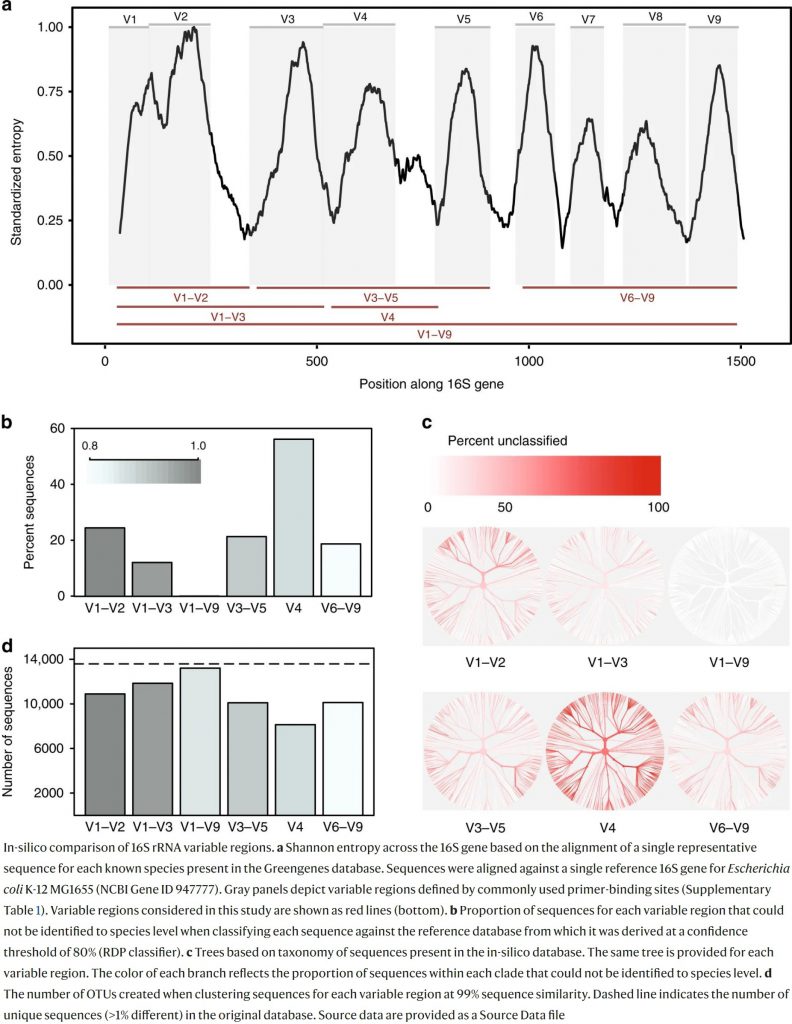
Q: What is the advantage of sequencing the full length of the 16S rRNA gene for targeted metagenomics?
A: Long reads are superior over short reads as they are able to cover the full-length 16S gene (V1 – V9) and are not limited to only the V3 – V4 partial gene region. Full length V1 – V9 16S metagenomics can provide comprehensive information on the microbial diversity compared to the short-read V3 – V4 partial region.
Q: How long will I get the results after I send my samples?
A: The TAT depends on the service committed.
General Sequencing Sample Requirements
- Minimum 5 ug and maximum 100 uL
- OD260/280 ratio > 1.8
- OD260/230 ratio > 2.0
- DNA quantification by using Qubit DNA Assay (recommended) or NanoDrop
- No degradation or RNA contamination
*May vary depending on sample type, sequencing service and platform
BTSeq™ Service
Sample requirements
- Minimum 500 ng (≥10 ng/ul) and maximum 50 uL of purified PCR product or plasmid
- OD260/280 ratio > 1.8
- OD260/230 ratio > 2.0
- DNA quantification using Qubit DNA Assay (recommended) or NanoDrop
- No degradation or RNA contamination
Publications
For inquiries, please share the following information with us:
- Project background/goals
- Genome/fragment size estimate
- Number of samples
- Type of sequencing (whole genome, RNA, amplicon, metagenomics etc)
- Preferred platform (Illumina, Pacbio, BTSeq etc) and specifications (PE150, PE250, CLR, HiFi etc)
- Sequencing depth/coverage
- Is bioinformatics needed? If yes, what type of analysis?
- Project timeline
- Budget estimate
